FastSurfer
Table of contents
Now that your images are in a suitable format we can use FastSurfer on them.
Do the following:
- Run FastSurfer on all your T1 scans using the XNAT Container Service
- Look at and understand the outputted results
What is FastSurfer?
We’ll be using FastSurfer, which is a deep-learning implementation of the FreeSurfer pipeline that allows us to conduct volumetric and surface-based analysis in a much shorter timeframe than FreeSurfer itself.
Running FastSurfer with the Container Service
This is where we start to see the real benefits of using the XNAT Container Service. Usually you would have to install and configure FreeSurfer yourselves and be limited to your local machine’s performance in running the processing. However we’re using a pre-set up container that is running on a GPU-enabled server - which means everything is already set to go and allows us to use FastSurfer with GPU support. This should allow us to perform volumetric analysis and surface-based thickness analysis in relatively short timeframes.
There is one additional pre-processing step that needs to be done. All the T1 images in the iBASH study cover a Field of View (FOV) that is greater than 256mm. They also have voxel resolutions of 1.1mm. FreeSurfer likes images to be 256x256x256 with 1mm isotropic resolition. So first we must conform the Nifti files to this.
To conform the Nifti into a FS-compatible input, you must run a container in a similar manner to dcm2niix:
- Go to your project
- Go to “Project Settings” in the “Actions” menu
- Go to “Container Service -> Configure Commands”
- Enable the “Converts NIFTI to 256 conformed MGZ file” command
- Go to an imaging session in your project
- Using the “Run” on the right hand side, convert any T1 NIfTI scan
- A dialog will pop up with runtime options. Please make sure
--cw256is entered into the MRICONVERTARGS field.
As with dcm2niix, you can monitor the processing from the “History” panel at the bottom of the imaging session page. Once complete, an MGZ (a file format often used by FreeSurfer) Resource will be created at the scan level.
Then, to run FastSurfer:
- Go to your project
- Go to “Project Settings” in the “Actions” menu
- Go to “Container Service -> Configure Commands”
- Enable the “Runs FastSurfer pipeline on a conformed T1 MGZ” command
- Go to an imaging session in the project that you wish to run FastSurfer on.
- In the action menu on the right hand side fo the session page, please select the Run Container’s action and click on the “Runs FastSurfer…” container.
- A dialog runtime box will come up. please make sure to enter
--parallelin the FASTSURF_ARGS field.
Interpreting Output
You can confirm that FastSurfer has successfully completed by looking at the bottom of the page of the session that you launched the container from. There should be one line in the history with Status of Complete and Name of “FastSurfer”. It would be helpful to look at the results from teh pipeline in order to make sure that FastSurfer produced reasonable output. There are a few ways of doing this:
- Go to “Manage Files” in the action menu of the right hand side of session page. It should open up a new box that provides a tree-like structure of the underlying file data.
- Uncheck everything except for the entry called FASTSURFER and click “Download” at the bottom of the window.
- Your browser will now download a zip file.
- Alternatively, you could script this download as you may have done in previous parts of this project.
- The easiest way to view the images would likely be via FreeView, FreeSurfer’s packaged image viewer, or fsleyes, the viewer from the FSL package. If either of those will be difficult to use, please let the course leads know.
- In the viewer, select the
nu.mgzfile from the mri subdirectory and theaparc.DKTatlas+aseg.deep.mgzfrom the same directory. - Temporarily turn off the second image (
aparc.DKTatlas+aseg.deep.mgz) so that you can first check that the volumetric T1 scan (nu.mgz) is of good quality and that there is limited motion or other artefact that might make the pipeline unreliable. From this scan, you can see that the cortical boundaries could be more distinct, but it should be alright.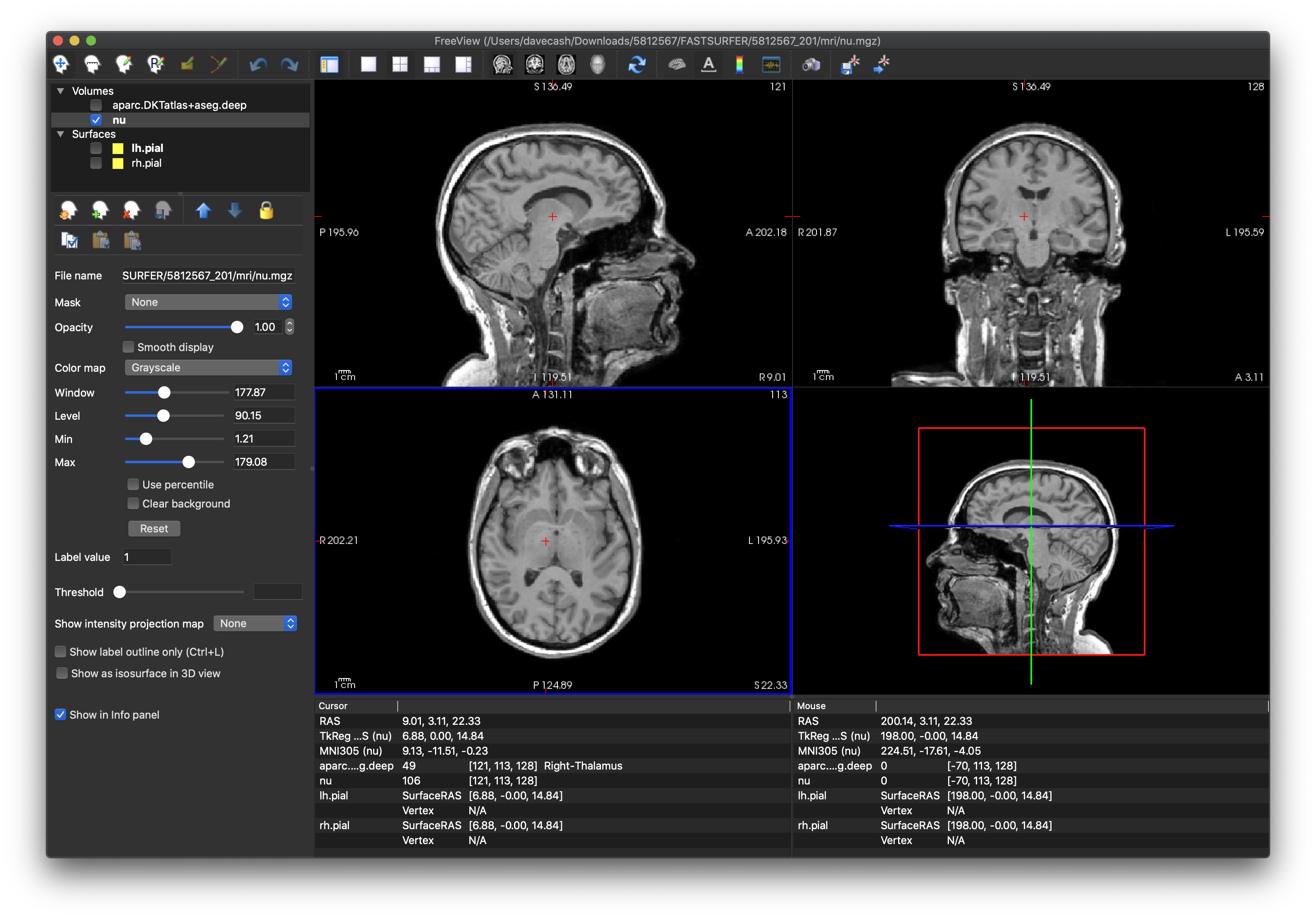
- Next, turn
aparc.DKTatlas+aseg.deep.withCC.mgzback on and select the colour map called “Lookup Table”, Make sure that the white matter is white and that the the coloured areas correspond to various parts of the cortical ribbon.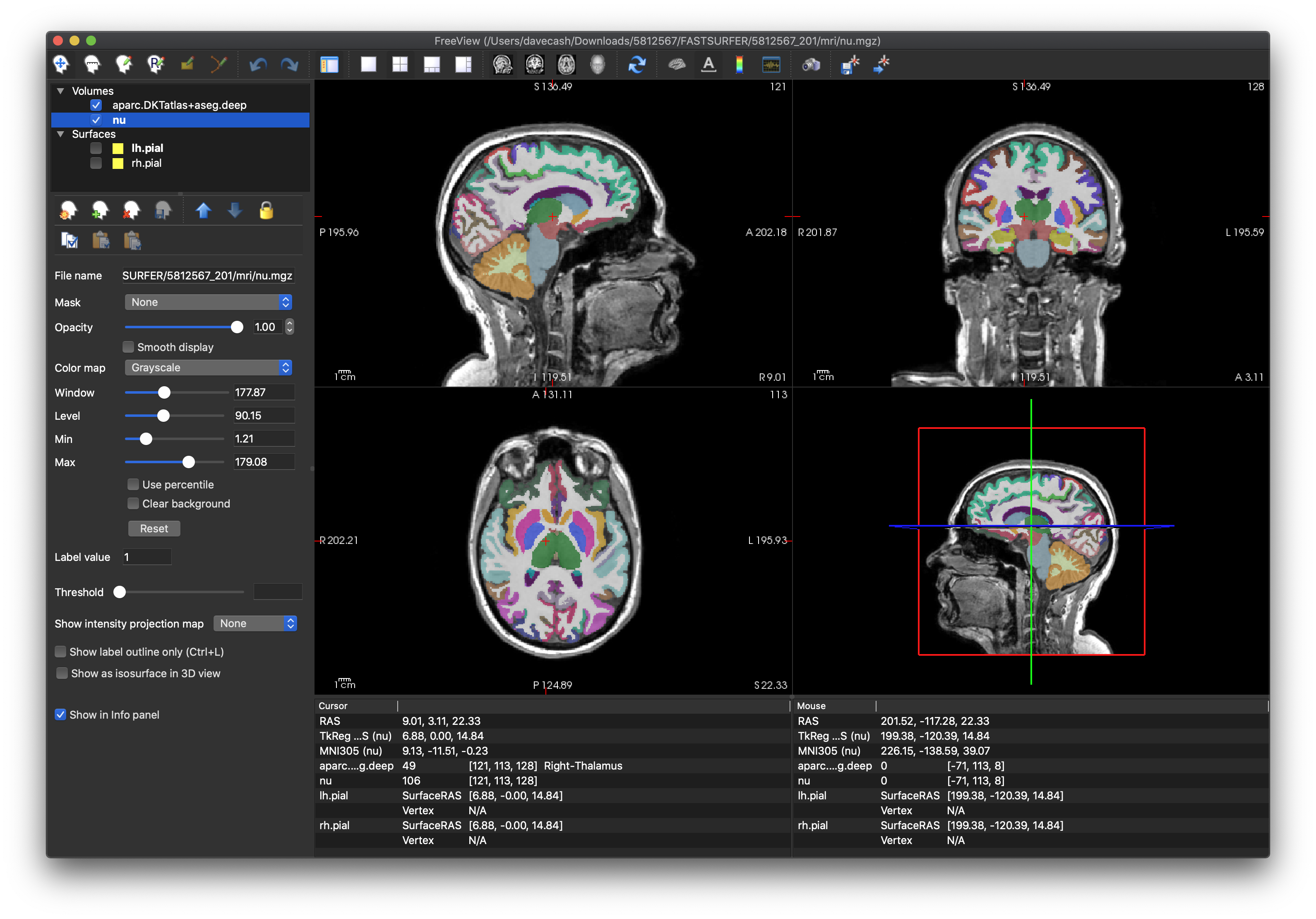
- Another way to review is to load the
lh.pialandrh.pialsurface files (from the surf subdirectory) and thenu.mgzimage volume again from above. The yellow curves representing the (left/right) hemisphere’s surface should closely adhere the boundary of the grey matter and the CSF.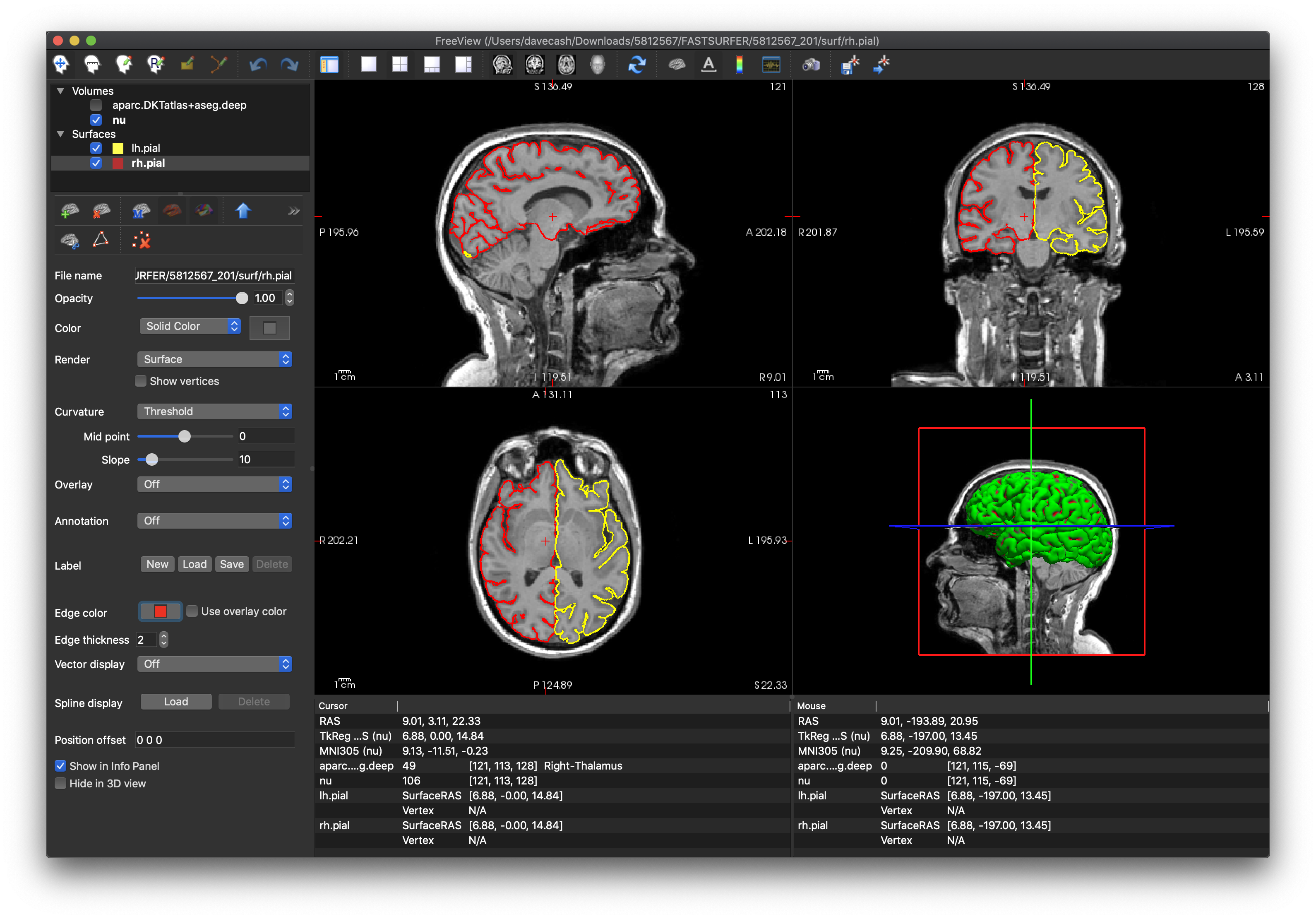
More detail of quality checking FastSurfer can be taken from this tutorial. There are also QA Tools available as part of this package if you would like to explore these as an option.
Once you have visually reviewed the data, the next step is to grab statistics. The file you want is in the stats subdiretory of the FASTSURFER resource, and it is called aseg+DKT.stats. The key part of the sample format is below.
# ColHeaders Index SegId NVoxels Volume_mm3 StructName normMean normStdDev normMin normMax normRange
1 2 277731 281844.1 Left-Cerebral-White-Matter 106.2308 6.4114 41.0000 131.0000 90.0000
2 4 13034 13776.2 Left-Lateral-Ventricle 37.4730 7.3857 22.0000 73.0000 51.0000
3 5 498 609.3 Left-Inf-Lat-Vent 56.8333 10.7469 31.0000 83.0000 52.0000
4 7 18989 19893.1 Left-Cerebellum-White-Matter 108.0617 7.6360 59.0000 129.0000 70.0000
5 8 65539 65687.7 Left-Cerebellum-Cortex 90.8048 13.9321 27.0000 126.0000 99.0000
6 10 6715 6360.5 Left-Thalamus-Proper 99.4649 10.2114 48.0000 118.0000 70.0000
7 11 2732 2697.0 Left-Caudate 86.3964 10.5292 54.0000 102.0000 48.0000
8 12 4736 4780.5 Left-Putamen 97.6617 4.2558 66.0000 108.0000 42.0000
9 13 1994 1948.2 Left-Pallidum 109.7849 3.7602 86.0000 124.0000 38.0000
Use your favorite tools to parse these text files and load them in your favorite software application to collate the data from multiple subjects and do some statistical analysis. You’ll look at these results again, once you have defaced the image in the next step.